We develop new AI tools to improve healthcare delivery, patient outcomes, and the overall management of healthcare systems.
A knowledge graph for clinical trials – CTKG

Abstract: Effective and successful clinical trials are essential in developing new drugs and advancing new treatments. However, clinical trials are very expensive and easy to fail. The high cost and low success rate of clinical trials motivate research on inferring knowledge from existing clinical trials in innovative ways for designing future clinical trials. In this manuscript, we present our efforts in constructing the first publicly available Clinical Trials Knowledge Graph, denoted as CTKG . CTKG includes nodes representing medical entities in clinical trials (e.g., studies, drugs and conditions), and edges representing the relations among these entities (e.g., drugs used in studies). Our embedding analysis demonstrates the potential utilities of CTKG in various applications such as drug repurposing and similarity search, among others.
Ziqi Chen, Bo Peng, Vassilis N. Ioannidis, Mufei Li, George Karypis, and Xia Ning. A knowledge graph for clinical trials – CTKG. Scientific Reports, 12(1):4724, February 2022. https://www.nature.com/articles/s41598-022-08454-z (CTKG is available in GitHub. A webportal is available here).
Understanding comorbidities and health disparities related to COVID-19: a comprehensive study of 776,936 cases and 1,362,545 controls in the state of Indiana, USA
 County-wise heatmap of Indiana displaying various epidemiological and socio-economic factors.
County-wise heatmap of Indiana displaying various epidemiological and socio-economic factors.
Abstract: Objective: To characterize COVID-19 patients in Indiana, United States, and to evaluate their demographics and comorbidities as risk factors to COVID-19 severity. Materials and Methods: EHR data of 776 936 COVID-19 cases and 1 362 545 controls were collected from the COVID-19 Research Data Commons (CoRDaCo) in Indiana. Data regarding county population and per capita income were obtained from the US Census Bureau. Statistical analysis was conducted to determine the association of demographic and clinical variables with COVID-19 severity. Predictive analysis was conducted to evaluate the predictive power of CoRDaCo EHR data in determining COVID-19 severity. Results: Chronic obstructive pulmonary disease, cardiovascular disease, and type 2 diabetes were found in 3.49%, 2.59%, and 4.76% of the COVID-19 patients, respectively. Such COVID-19 patients have significantly higher ICU admission rates of 10.23%, 14.33%, and 11.11%, respectively, compared to the entire COVID-19 patient population (1.94%). Furthermore, patients with these comorbidities have significantly higher mortality rates compared to the entire COVID-19 patient population. Health disparity analysis suggests potential health disparities among counties in Indiana. Predictive analysis achieved F1-scores of 0.8011 and 0.7072 for classifying COVID-19 cases versus controls and ICU versus non-ICU cases, respectively. Discussion: Black population in Indiana was more adversely affected by COVID-19 than the White population. This is consistent to findings from existing studies. Our findings also indicate other health disparities in terms of demographic and economic factors. Conclusion: This study characterizes the relationship between comorbidities and COVID-19 outcomes with respect to ICU admission across a large COVID-19 patient population in Indiana.
Nader Zidan, Vishal Dey, Katie Allen, John Price, Sarah Renee Zappone, Courtney Hebert, Titus Schleyer, and Xia Ning. Comorbidities and socio-economic factors affecting COVID-19 severity: A comprehensive study of 776,936 cases and 1,362,545 controls in the State of Indiana, USA. JAMIA Open, 6(1):ooad002, 02 2023. https://academic.oup.com/jamiaopen/article/6/1/ooad002/7024622
A multi-layered GRU model for COVID-19 patient representation and phenotyping from large-scale EHR data
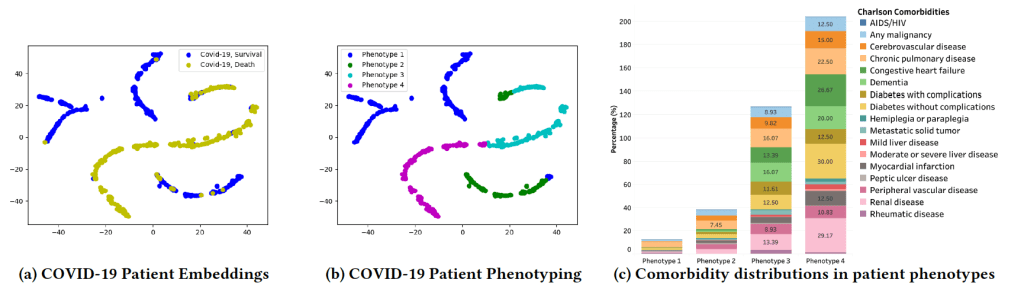
Abstract: The unprecedented scale of the COVID-19 pandemic created an alarming shortage of healthcare resources. To enable a more efficient resource allocation and targeted treatment, in this manuscript, we conducted a data-driven study of COVID-19 patients to predict patient outcomes and identify patient phenotypes. Specifically, we developed a multi-layered gated recurrent units-based model, referred to as mGRU-CP, to learn patient embeddings and estimate patient survival probabilities by leveraging their electronic health record (EHR) data in the COVID-19 Research Data Commons. We empirically compared mGRU-CP against four state-of-the-art baseline methods on three sets of patient features. The experimental results demonstrate that mGRU-CP could achieve competitive or superior performance over the baseline methods in all the settings. Our analysis also shows that the learned patient embeddings in mGRU-CP could enable meaningful patient phenotyping to better understand patient mortalities. Our study is significant in understanding patients in the past COVID-19 pandemic, and provides computational tools to predict patient outcomes and inform associated healthcare resource allocation for the future pandemics proactively.
Arpita Saha, Maggie Samaan, Bo Peng◦, and Xia Ning. A multi-layered gru model for covid-19 patient representation and phenotyping from large-scale ehr data. In Proceedings of the 14th ACM International Conference on Bioinformatics, Computational Biology, and Health Informatics, BCB ’23, pages 1–6, New York, NY, USA, 2023. Association for Computing Machinery. https://dl.acm.org/doi/10.1145/3584371.3612986
Hybrid collaborative filtering methods for recommending search terms to clinicians

Abstract: With increasing and extensive use of electronic health records (EHR), clinicians are often challenged in retrieving relevant patient information efficiently and effectively to arrive at a diagnosis. While using the search function built into an EHR can be more useful than browsing in a voluminous patient record, it is cumbersome and repetitive to search for the same or similar information on similar patients. To address this challenge, there is a critical need to build effective recommender systems that can recommend search terms to clinicians accurately. In this study, we developed a hybrid collaborative filtering model to recommend search terms for a specific patient to a clinician. The model draws on information from patients’ clinical encounters and the searches that were performed during them. To generate recommendations, the model uses search terms which are (1) frequently co-occurring with the ICD codes recorded for the patient and (2) highly relevant to the most recent search terms. In one variation of the model (Hybrid Collaborative Filtering Method for Healthcare, or HCFMH), we use only the most recent ICD codes assigned to the patient, and in the other (Co-occurrence Pattern based HCFMH, or cpHCFMH), all ICD codes. We have conducted comprehensive experiments to evaluate the proposed model. These experiments demonstrate that our model outperforms state-of-the-art baseline methods for top-N search term recommendation on different data sets.
Zhiyun Ren, Bo Peng, Titus Schleyer, and Xia Ning. Hybrid collaborative filtering methods for recommend- ing search terms to clinicians. Journal of Biomedical Informatics, 113:103635, December 2020. https://www.sciencedirect.com/science/article/pii/S153204642030263X?via%3Dihub#f0030