As the most abundant biological entities in aquatic environments, viruses have significant influences on microbial mortality, genetic diversity, and biogeochemical cycling that we are just beginning to fully appreciate. One limitation to our investigation of marine viruses has been a lack of quantitative methods to explore their diversity, ecology, evolution, and interactions with their hosts. At the Sullivan Lab, we are developing new methods and techniques to explore viral roles in nature. To this end, we have evaluated existing and new methods for concentrating, purifying, and sequencing ocean viral DNA to develop a quantitative sample-to-sequence metagenomic pipeline for ocean viruses. We have also explored new ways to link viruses to their hosts either one cell at a time (e.g., phageFISH) or en masse at a population scale (e.g., viral tagging). In additon, we have developed methods to quantitatively characterize viral morphology in complex ocean communities (quantitative transmission electron microscopy, qTEM) and to bring annotation to unknown viral sequence space through experimentally determining virion structural proteins in isolates and in nature using (meta)proteomics.
For bioinformatics development, please visit our software page.
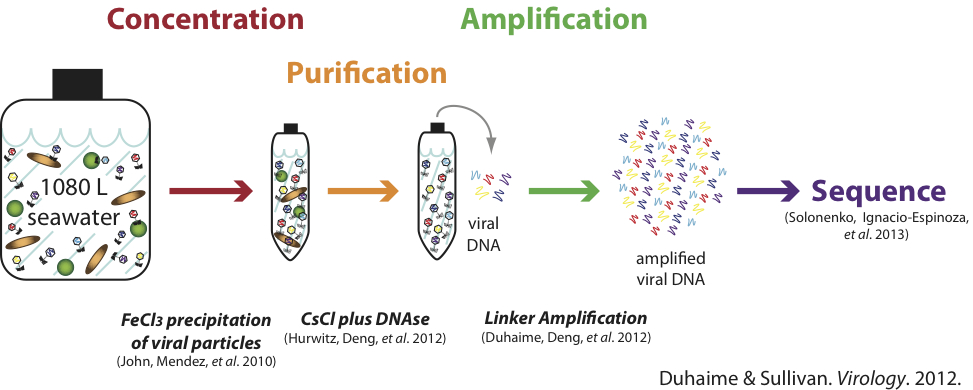
Sample-to-Sequence Pipeline
Purpose: A pipeline to generate replicable, near-quantitative metagenomic datasets from ultra-low dsDNA samples including natural viral communities.
Brief Description: Viruses from <0.2 µm seawater samples are concentrated at high efficiency using FeCl3 precipitation, purified with CsCl density gradients and DNase, then amplified with low-bias linker amplification.
References:
- Duhaime, M.B., & Sullivan, M.B. (2012). Ocean viruses: Rigorously evaluating the metagenomic sample-to-sequence pipeline.Virology. 434(2), 181-186. doi:10.1016/j.virol.2012.09.036. LINK
- John, S.G., Mendez, C.B., Deng, L., Poulos, B., Kauffman, A.K.M., Kern, S., Brum, J., Polz, M.F., Boyle, E.A., & Sullivan, M.B. (2011). A simple and efficient method for concentration of ocean viruses by chemical flocculation. Environ Microbiol Rep.3(2), 195-202. doi:10.1111/j.1758-2229.2010.00208.x. LINK
- Duhaime, M.B., Deng, L., Poulos, B.T., & Sullivan, M.B. (2012). Towards quantitative metagenomics of wild viruses and other ultra-low concentration DNA samples: A rigorous assessment and optimization of the linker amplification method. Environ Microbiol. 14(9), 2526-2537. doi:10.1111/j.1462-2920.2012.02791.x. LINK
- Hurwitz, B.L., Deng, L., Poulos, B.T., & Sullivan, M.B. (2012). Evaluation of methods to concentrate and purify ocean virus communities through comparative, replicated metagenomics. Environ Microbiol. 15(5), 1428–1440. doi:10.1111/j.1462-2920.2012.02836.x. LINK
- Solonenko, S.A., Ignacio-Espinoza, J.C., Alberti, A., Cruaud, C., Hallam, S., Konstantinidis, K., Tyson, G., Wincker, P., &Sullivan, M.B. (2013). Sequencing platform and library preparation choices impact viral metagenomes. BMC Genomics. 14(1), 320. doi:10.1186/1471-2164-14-320. LINK
phageFISH
Left: Use of phageFISH to visualize specific phages and hosts in a mixed population.
Right: Epifluorescence micrograph of viruses (PSA-HP1; red fluorescence) infecting their host (Pseudoalteromonas sp. H100; green fluorescence).
Purpose: Detect and visualize intra-and extracellular phage DNA to elucidate phage-host interactions in both field and laboratory studies, including investigation of temperate phages and systems where genetic manipulation is not feasible.
Brief Description: An improved geneFISH protocol (>92% efficiency) to detect and quantify viral genes using epifluorescence microscopy.
Reference:
- Allers, E.*, Moraru, C.*, Duhaime, M.B., Beneze, E., Solonenko, N., Barrero-Canosa, J., Amann, R.†, & Sullivan, M.B.†(2013). Single-cell and population level viral infection dynamics revealed by phageFISH, a method to visualize intracellular and free viruses. Environ Microbiol. 15(8), 2306–2318. doi:10.1111/1462-2920.12100. (* =co-first authors) († =co-corresponding authors) LINK
- Textbook chapter: PhageFISH for Monitoring Phage Infections at Single Cell Level: LINK
Viral Tagging
Left: (A) Fluorescent labeling of viruses and attachment to their hosts (Synechococcus sp. WH7803). (B) Flow-cytometry plot showing differential fluorescence signals of host cells that are or are not tagged with an infecting virus, with (C) corresponding micrographs and (D) PCR results showing detection of viruses.
Right: Population genome landscape plot showing the genetic relationship (average amino acid identity) of cultivated and viral-tagged T4-like phages of Synechococcus WH7803 from a single seawater sample, as well as all available marine cyanophage genomes.
Purpose: A high-throughput method to investigate viruses that infect a specific microbial host to understand viral-host interactions and natural host-specific viral diversity and evolution.
Brief Description: Natural viral communities are fluorescently stained with the nucleic acid stain SYBR Green I, incubated with a cultivated microbial host, and then sorted with a flow-cytometer to obtain the viral-tagged cells based on their differential fluorescence. Metagenomic sequencing of the sorted samples then enables genomic examination of viruses that infect the particular host.
References:
- Deng, L., Gregory, A., Yilmaz, S., Poulos, B.T., Hugenholtz, P., & Sullivan, M.B. (2012). Contrasting life strategies of viruses that infect photo- and heterotrophic bacteria, as revealed by viral tagging. mBio. 3(6), e00373-12. doi:10.1128/mBio.00373-12. LINK
- Deng, L.*, Ignacio-Espinoza, J.C.*, Gregory, A., Poulos, B.T., Weitz, J.S., Hugenholtz, P., & Sullivan, M.B. (2014). Viral tagging reveals discrete populations in Synechococcus viral genome sequence space. Nature. doi:10.1038/nature13459. (*=co-first authors) LINK
qTEM (Quantitative Transmission Electron Microscopy)
Left: Average percentage of viral morphotypes in 26 global ocean samples collected on the Tara Oceans Expedition. Non-tailed viruses dominated these global ocean samples generating a new paradigm of non-tailed viral dominance in marine environments.
Right: Correspondence analysis comparing global ocean viral communities based on the distribution of viral capsid diameters. Vectors correspond to the influence of environmental variables, with salinity, oxygen, and temperature as significant influences.
Purpose: Quantitatively determine morphological characteristics of aquatic viral assemblages to assess viral morphotype composition and distribution of viral capsid widths and tail lengths.
Brief Description: Viruses in aquatic samples are quantitatively deposited onto grids using an air-driven ultracentrifuge, positively stained with uranyl acetate, and examined using transmission electron microscopy.
Reference:
- Brum, J.R., Schenck, R.O., & Sullivan, M.B. (2013). Global morphological analysis of marine viruses shows minimal regional variation and dominance of non-tailed viruses. ISME J. 7, 1738–1751. doi:10.1038/ismej.2013.67. LINK
Targeted Viral Metagenomics
Purpose: Investigate viral genomes with specific genes of interest (e.g., metabolic genes), or viruses with specific morphotypes (e.g., non-tailed viruses).
Brief Description: Natural viral communities are separated based on buoyant density and surface charge, decreasing viral diversity with each fractionation step. Fractions enriched with viruses containing specific genes of interest (detected by pcr) are then sequenced and assembled to investigate the target gene within genomic context. Viruses with specific morphotypes can also be targeted by screening each fraction using qTEM.
References: Based on the following:
- Brum, J.R., Culley, A.I., and Steward, G.F. (2013) Assembly of a marine viral metagenome after physical fractionation. PLoS ONE 8:e60604
- Brum, J.R., and Steward, G.F. (2011) Physical fractionation of aquatic viral assemblages. Limnology and Oceanography: Methods 9:150