Oceans Research in the Sullivan Lab
The oceans cover ~2/3 of our planet, halve the carbon dioxide greenhouse gases humans release into the atmosphere, and provide food for billions of people. Microbes are central to these ecosystem services as they are abundant and drive many biogeochemical cycles, though they do so under constraints imposed by viruses. We investigate the roles of marine viruses in regulating microbial communities through mortality, horizontal gene transfer, and metabolic reprogramming. Our field work ranges from local temporal studies (e.g., LineP, Line67, BATS) to global oceanographic surveys (e.g., Tara Oceans), as well as from surface waters anywhere (including the Arctic and the Antarctic) to the deepest trenches, remote coral reefs, and diverse bottom waters. Separately, these data-driven discovery efforts are complemented by hypothesis testing, particularly through development and multi-omics study of new phage-host model systems (e.g., Bacteroidetes and Pseudoalteromonas). This long-standing research focus in the group has helped establish the experimental and informatic quantitative viral ecogenomic approaches that are transforming our understanding of viruses that infect microbes in complex communities.
Key viral ecogenomic research Qs include:
- What are biologically meaningful units for viruses (genus, species, sub-species)?
- What are the patterns and drivers of viral community structure?
- What roles do viruses play in ocean ecosystems?
As a coordinator and steering commitee member of the Tara Oceans Consortium, the Sullivan Lab is at the forefront of developing Ecosystems Biology approaches to studying viruses, microbes and eukaryotes in complex systems – in collaboration with dozens of labs across as many countries. This work has led to several paradigm shifting advances – including that viruses shunt and shuttle carbon and best predict carbon flux to the deep sea, as well as that symbioses dominate pelagic water plankton – and continues to provide a rich opportunity for discoveries in the Sullivan Lab. Our current work is focused on understanding microdiversity in dsDNA viruses, the fraction of microbial metabolic pathways that viruses directly manipulate, establishing baseline ecological data for RNA viruses in the global oceans, and assessing informatic, experimental and theoretical means to infer virus-host interactions. This work is funded by the Gordon and Betty Moore Foundation and the NSF, as well as DOE EMSL User Awards.
Navigate to:
– Oceans research feature
– Recent publications in oceans research
– Oceanic datasets
– Lab members involved in ocean research
– Field work in oceanic environments
– TARA expeditions information
– Oceanic model systems
Oceans Research Features
Phosphate stress:
Phosphate is a key limiting nutrient in the oceans, yet very little is known about the effects of phosphate limitation in oceanic bacteria. Our research focuses on developing a model system to explore the impact of phosphate limitation in Pseudoalteromonas, a heterotrophic bacterium commonly found in the oceans. We are characterizing the phosphate stress response by measuring the expression of genes and proteins during phosphate stress. We will then use these conditions to answer the question: how does phosphate limitation affect phage infection?
Marine arctic viral communities:

Recent Oceans Publications
- Zimmerman, A., Howard-Varona, C., Needham, D.M., John, S., Worden, A.Z., Sullivan, M.B., Waldbauer, J.R., & Coleman, M.L. 2019. Metabolic and biogeochemical consequences of viral infection in aquatic ecosystems. Nature Reviews Microbiology. 18:21-34. LINK PDF
- Zimmerman, A.E., Bachy, C., Ma, X., Roux, S., Jang, H.B., Sullivan, M.B., Waldbauer, J.R., & Worden, A.Z. 2019. Closely related viruses of the marine picoeukaryotic alga Ostreococcus lucimarinus exhibit different ecological strategies. Environmental Microbiology. 21(6):2148–2170. LINK PDF
- Ibarbalz, F.M., Henry, N., Brandao, M.C., Martini, S., Busseni, G., Byrne, H., Coelho, L.P., Endo, H., Gasol, J.M., Gregory, A.C., Mahe, F., Rigonato, J., Royo-Llonch, M., Salazar, G., Sanz-Saez, I., Scalco, E., Soviadan, D., Zayed, A.A., Zingone, A., Labadie, K., Ferland, J., Marec, C., Kandels, S., Picheral, M., Dimier, C., Poulain, J., Pisarev, S., Carmichael, M., Pesant, S., Tara Oceans Coordinators, Babin, M., Boss, E., Iudicone, D., Jaillon, O., Acinas, S.G., Ogata, H., Pelletier, E., Stemmann, L., Sullivan, M.B., Sunagawa, S., Bopp, L., de Vargas, C., Karp-Boss, L., Wincker, P., Lombard, F., Bowler, C., & Zinger, L. 2019. Global Trends in Marine Plankton Diversity across Kingdoms of Life. Cell. 179:1084-1097. LINK PDF
- Salazar, G., Paoli, L., Alberti, A., Huerta-Cepas, J., Ruscheweyh, H.J., Cuenca, M., Field, C.M., Coelho, L.P., Cruaud, C., Engelen, S., Gregory, A.C., Labadie, K., Marec, C., Pelletier, E., Royo-Llonch, M., Roux, S., Sánchez, P., Uehara, H., Zayed, A.A., Zeller, G., Carmichael, M., Dimier, C., Ferland, J., Kandels, S., Picheral, M., Pisarev, S., Poulain, J., Tara Oceans Coordinators, Acinas, S.G., Babin, M., Bork, P., Bowler, C., de Vargas, C., Guidi, L., Hingamp, P., Iudicone, D., Karp-Boss, L., Karsenti, E., Ogata, H., Pesant, S., Speich, S., Sullivan, M.B., Wincker, P., & Sunagawa, S. 2019. Gene Expression Changes and Community Turnover Differentially Shape the Global Ocean Metatranscriptome. Cell. 179:1068-1083. LINK PDF
Oceans Datasets
Oceans Researchers
- Dean Vik
- ZhiPing Zhong
- Cristina Howard-Varona
- Jingjie Du
- Guillermo Dominguez Huerta
- Ahmed Zayed
- Funing Tian
- Marissa Gittrich
- Lindsey Solden
- Christine Sun
- James Wainaina
- Dylan Cronin
- Sergei Solonenko
- Ho Bin Jang
Global Oceanographic Research Expeditions
The Sullivan Lab collaborates with several expeditions to obtain diverse samples to answer important biological questions.
The Sullivan Lab is amassing an unprecedented archive of systematically-collected, metadata-rich ocean viral concentrates from two European-funded global oceanographic research expeditions, Tara Oceans and Malaspina. In total, we anticipate > 2,000 samples from multiple depths at 550 sites around the world’s oceans. Both expeditions explore the biology, chemistry, and physics of the oceans through large, international consortiums that include unprecedentedly diverse expertise. Further, both consortiums are built upon open exchange of ideas, well–documented voucher collections, and robust databasing to support sample archives. This foundation for diversity studies on a scale not previously possible allows us to explore the diversity, phylogeography, and genomic variability of ocean viral groups.
Tara Oceans
Tara Expeditions, a French non–profit organization active since 2003, organizes voyages to study the world’s oceans and the impact of climate change. Tara, a boat built for extreme conditions, is the platform for high–level scientific research missions around the globe.
Tara Oceans, a three year expedition that took place from September 2009 to November 2012, was the first attempt at a global study of marine plankton (viruses, bacteria, protists, and small metazoans). This expedition consisted of 375 stations sampling at 1 to 5 depths per station, to 1000m, mostly of the photic zone. Samples and data collected from across the world’s ocean environments are being analyzed by a scientific consortium of about 200 scientists with participation from 50 labs in 15 countries (including the Tucson Marine Phage Lab) to gain better knowledge of marine ecosystems. What do communities look like? How do various organisms interact with each other? How do they interact with their physico-chemical environment?
 Tara Oceans Special Issue in Science
Tara Oceans Special Issue in Science
In a special issue of Science, five papers uncover new information about the ways in which marine viruses and microbes interact on a global scale. The work is part of the Tara Oceans Expedition, a global effort to understand complex interactions among ocean ecosystems, climate, and biodiversity.
Malaspina
Malaspina Expedition 2010 is an interdisciplinary research project led by the Spanish National Research Council took place from December 2010 to July 2011. This expedition, aboard the ship Hesperides, voyaged through Rio de Janeiro, Cape Town, Perth, Sydney, Auckland, Honolulu, Cartagena of Indias, and Panama before returning to Spain. It sampled at 180 stations with 9 depths per station, to 5000m, of the photic zone and deep sea. About 300 scientists are directly involved with participation from 30 labs in 14 countries (including Tucson Marine Phage Lab). The goal of this expedition is to generate an inventory of the impact of global change on the ecosystems of the ocean and to explore their biodiversity, particularly in the deep ocean.
• Malaspina Expedition Website
Coastal to Open Ocean Transects
Line P
The LineP cruises from Vancouver to Ocean Station Papa traverse some 2000km along the climate-sensitive Oxygen Minimum Zone (OMZ) of the subarctic Eastern Northern Pacific Ocean. The subarctic Eastern North Pacific (ENP) Ocean hosts a massive, deep-water OMZ and large annual open-ocean dimethylsulfide (DMS) pulses. Marine OMZs are sources for climatologically active trace gases including methane, nitrous oxide, and dimethylsulfide, and major sinks for nitrogen. Microbially–mediated biological activity within these systems thus directly impacts both ocean productivity and global climate balance. In addition, ENP surface waters see large annual blooms of phytoplankton and bloom-associated bacteria that generate significant fluxes of DMS, which when “leaked” to the atmosphere influences global climate through cloud formation. The LineP transect of the ENP OMZ has been oceanographically monitored for >50 years, and in recent years has adopted a microbiological research focus.
Our research seeks to explore the roles that viruses play in these systems, in the context of Dr. Steve Hallam Lab’s microbial prokaryote work. What types of viruses are there, and how are they distributed throughout 4,000m of the water column? What microbial metabolisms might they be manipulating (e.g., photosynthesis)? How do their communities change over space and time?
Line 67
The Line67 transect was initiated ∼1950 as part of the CalCOFI program and is a designated time series site with over 20 years of contextual data maintained between MBARI and Scripps Institution of Oceanography. This coastal to open ocean transect traverses a Monterey Bay coastal area influenced by agricultural runoff, natural mesotrophic waters, and oligotrophic waters all through one of the most well–known and well–studied upwelling regions in the world, the Monterey Bay.
Our research focuses on studying how viral communities change along this oceanographically rich transect, as well as how viruses impact their microbial prokaryote and picoeukaryote hosts. This work is in collaboration with Dr. Alex Worden, MBARI.
Time Series
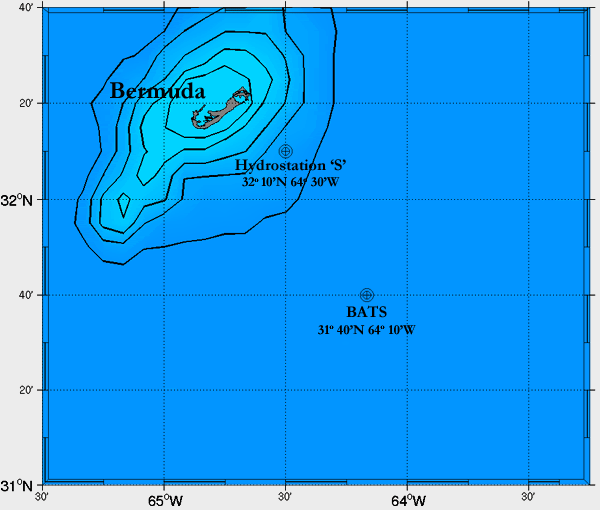
BATS (Bermuda Atlantic Time-series Study)
The Bermuda Atlantic Time-series Study (BATS) off the coast of Bermuda was established in 1988 to allow scientists to monitor data on dissolved carbon in the ocean and carbon dioxide in the atmosphere. This data helps scientists better understand global climate change and the ocean’s responses to variations in the Earth’s atmosphere, and can be used to study many ocean dynamics such as temporal variations in nutrient concentrations of surface water, phytoplankton blooms, the role of dissolved organic carbon in the global carbon cycle, and responses of calcifying organisms (like corals) to ocean acidification.
Here at the Sullivan Lab, we are partipating in the BATS sampling expeditions to understand how nutrient availability affects viral dynamics, the contributions of viral genes to nutrient uptake by the hosts during infection, and the chemical composition of nutrients released from host cells during viral infection.
Oceanic Model Systems
At the Sullivan Lab, we have been developing new phage-host model systems with phages infecting heterotrophic bacteria (heterophages), as well as advancing more well-established model systems with phages infecting marine cyanobacteria (cyanophages).
Cyanophages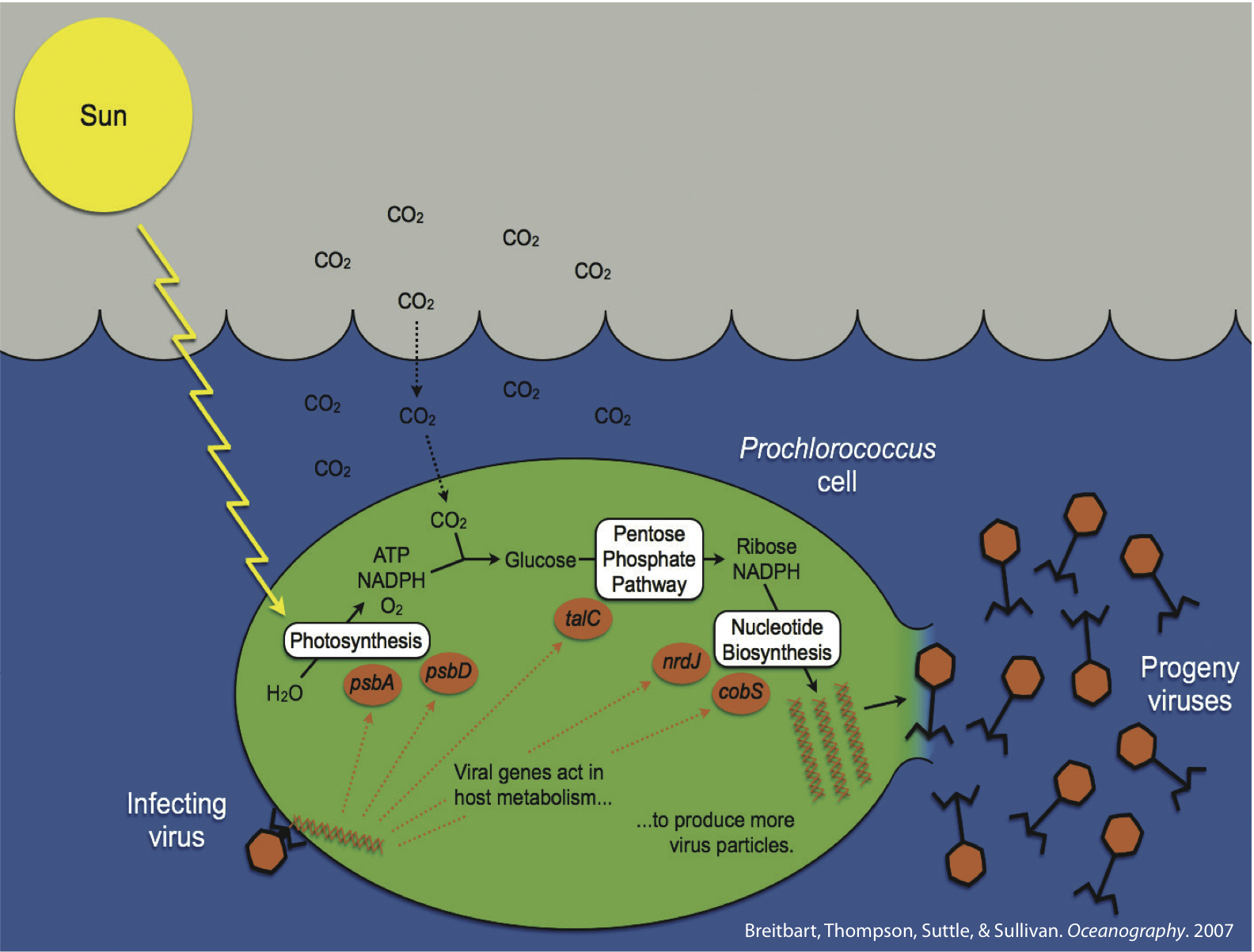
The marine cyanobacteria Prochlorococcus and Synechococcus are globally important primary producers. In spite of their small size, these cyanobacterial cells are numerically dominant over vast areas of the “desert oceans” and are significant contributors to global carbon cycling. These were some of the earliest “ecological microbes” sequenced by the DOE JGI, which led to an understanding of the genomic underpinnings that are responsible for their ecological niches in the environment
Cyanobacterial viruses (cyanophages) are abundant, contribute to host mortality, and impact marine cyanobacterial diversity through mortality and moving genes through the host population. Cyanophage genomes appear to be standard “coliphage” decorated with a suite of niche- and host-defining genes. These latter genes include photosynthesis genes, as well as genes likely involved in carbon metabolism, phosphate stress and novel nucleotide metabolism genes. Detailed studies of the core reaction center genes suggest that they are widespread among cyanophage isolates with a seemingly predictable distribution, they are expressed during infection, and parts of the viral copies of the genes have even been transferred back into their hosts – thus cyanophages are acting as evolutionary drivers of the core reaction centers of the numerically dominant photosystems on the planet.
Heterophages
We are interested in understanding diverse, environmental heterotrophic phages, and for that we isolate and study new heterotrophic phage-host model systems. We mechanistically explore all biological aspects of phage-host interactions, including adsorption, host takeover, phage transcription, DNA replication, phage virion formation, and cell lysis. For this we use a suite of experimental and informatics technologies that encompass traditional phage work, microscopy (including electron microscopy and phageFISH), genomics, transcriptomics, proteomics and various metabolomics approaches.