(revised – August 2021)
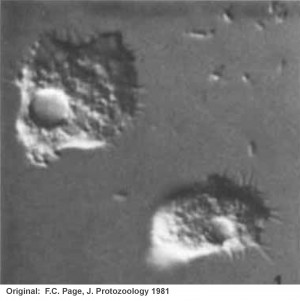
The genus Protacanthamoeba was first described by Page (1981) on the basis of microscopic analysis of morphology. (See Figures to left from Page, 1981; upper figure is of Protacanthamoeba caledonica trophozoite; lower figure is of cysts from same isolate).
In his description, the genus Protacanthamoeba included the type-species P. caledonica, for which strain 229 was isolated from salt water of the Morar estuary in western Scotland.
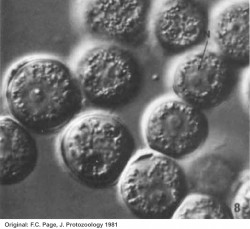 Page found that Protacanthamoeba differed from Acanthamoeba in the structure of the cyst. He noted that although the amoebae he isolated had a trophic form very much like that of Acanthamoeba, the cyst was without the opercula and preformed points of excystment characteristic of Acanthamoeba. In the locomotive form the amoebae was essentially indistinguishable from Acanthamoeba.
Page found that Protacanthamoeba differed from Acanthamoeba in the structure of the cyst. He noted that although the amoebae he isolated had a trophic form very much like that of Acanthamoeba, the cyst was without the opercula and preformed points of excystment characteristic of Acanthamoeba. In the locomotive form the amoebae was essentially indistinguishable from Acanthamoeba.
Isolates attributed to Protacanthamoeba were only rarely reported following the formal description by Page. This has changed slightly following the application of molecular approaches to identify isolates. Nevertheless, isolates of Protacanthamoeba are more rarely reported than most of the other FLA considered on this site.
MOLECULAR STUDIES OF PROTACANTHAMOEBA
The first sequence of Protacanthamoeba, representing the newly described species P. bohemica, was produced by Vykova et al. in 2005. This is the only sequence attributed directly to Protacanthamoeba that can be considered to be complete or nearly complete, with a length of 2028 bp. (The next longest sequence is an unpublished partial sequence of the P. caledonica Page isolate (CCAP 1567/1; ATCC 30976) that our lab at OSU has obtained, that differs from P. bohemica by 1.4%.) Although slightly longer than the “normal” eukaryotic nuclear small subunit (18S) rRNA gene, the sequence of Protacanthamoeba differs from those found in isolates of Acanthamoeba by the lack of the hypervariable expansion regions of the gene that characterize the 18S rRNA gene of all acanthamoebae. As of July 2018, 34 sequences of the 18S rRNA gene had been deposited (or were available to us) under the generic name “Protacanthamoeba”
In addition to these sequences, 31 other sequences exist in the DNA databases that we attribute to taxa within the general description of Protacanthamoeba. Our criterion for attributing a sequence to Protacanthamoeba is that sequence similarity is greater than 91% to either the P. bohemica or P. caledonica sequences. This compares to a general sequence similarity of ~85% to sequences from isolates of Acanthamoeba. All but one of the 20 additional sequences are less than 1000 bp in length. The uncultured eukaryote gene clone AD_Mclone16 shows 92% sequence similarity to the sequence of P. bohemica, and 93% similar to the OSU sequence of P. caledonica. (P. bohemica and P. caledonica were found to be 98% similar).
The distribution of sequence lengths for the sequences as of January 2018 is shown in the accompanying figure.
Examination of the sequence similarities among the 65 available sequences that we attribute to Protoacanthamoeba suggests that the genus may represent a fairly diverse group of isolates. How much species structure exists in the genus is unclear, although there is a suggestion of at least two or three differentiated subgroups.
Careful phylogenetic analysis is difficult for several reasons. First, analysis is restricted by the shorter length of the incomplete sequences for isolates of Protacanthamoeba (compared to almost complete sequences from Acanthamoeba or Vermamoeba). Second, because of small size and different approaches to obtain sequences of Protacanthamoeba, there is a lack of major overlap between all sequences. In fact, there is only a single 314 base region in which 39 of the 65 sequences overlap. There is no segment shared by all of the sequences. Construction of a phylogenetic tree relating the isolates is difficult because of the lack of sufficient overlap of sequences. Comparisons of pairwise sequence differences yields values in the range 0 – 0.084. More than half of pairwise comparisons yield sequence differences that are less than 1%, while only 13% of the pairwise comparisons yield sequence differences that exceed 7%.
ISOLATES IDENTIFIED IN ENVIRONMENTAL SCREENS
Of the 65 DNA sequences attributed to Protacanthamoeba that are represented in the figure above, 30 are identified in the DNA databases as uncultured eukaryotic sequences (46%), a frequency much greater than seen for Acanthamoeba, Balamuthia or Naegleria, but a lower frequency than was seen for Vermamoeba. Nonetheless, the overall frequency with which sequences attributed to Protacanthamoeba are observed in random environmental studies is low.
Additional Protacanthamoeba sequences are appearing in large environmental surveys based on new NextGen sequencing methods and deposited in the databases as Sequence Read Archives (SRA). As we screen the data of SRA’s we will report and add this data to our information about Protacanthameoba.