(updated December 2017)
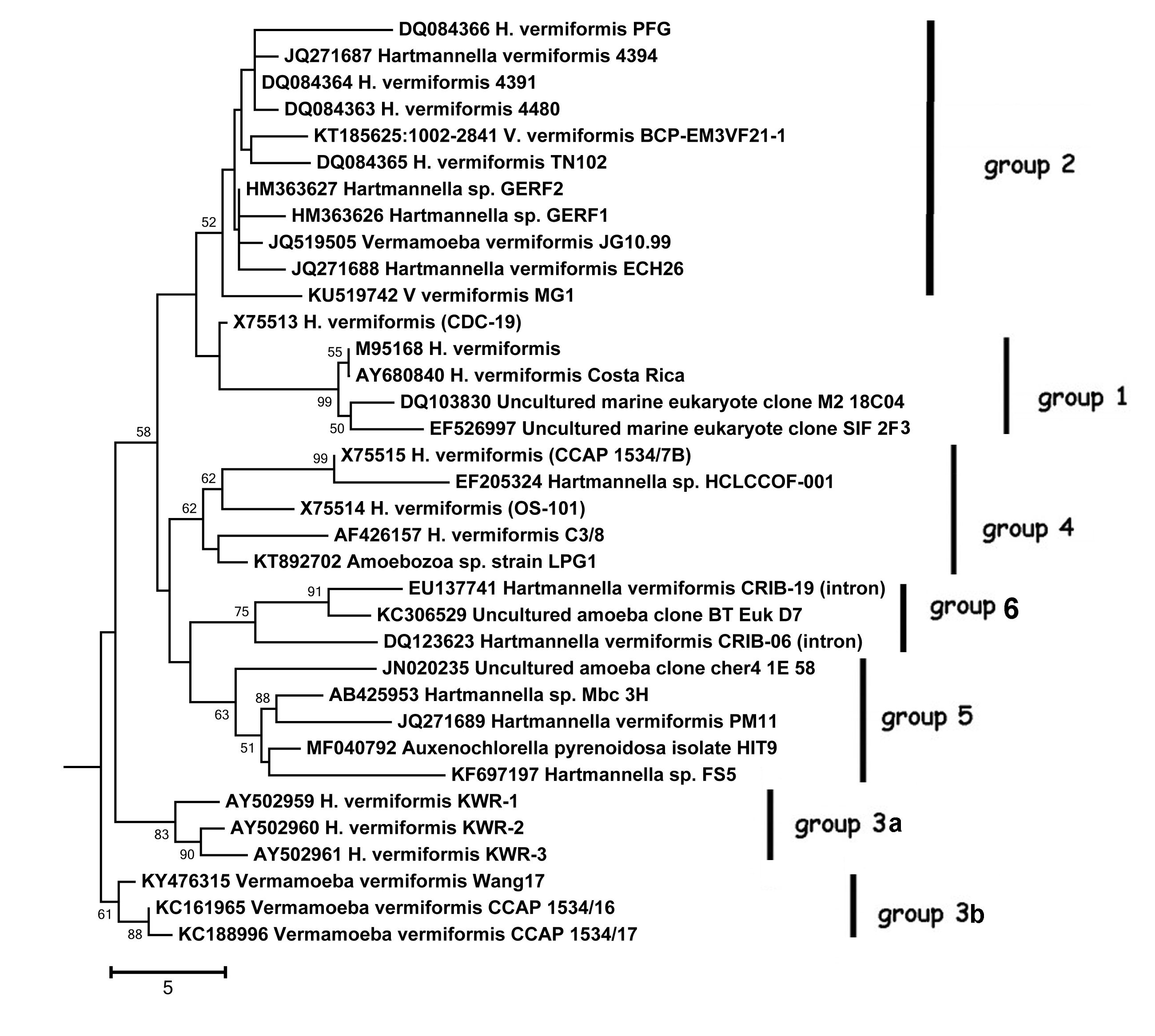
Phylogenetic relationships have been examined for 35 isolates of V. vermiformis in which sequences for the 18S rRNA gene span all or most of the gene.
The average pairwise sequence similarity among these isolates was 0.9923, while the greatest pairwise divergence was only 0.0204. In the context of variation within Acanthamoeba, isolates of V. vermiformis show much less divergence. The level of variation is less than that which occurs within sequence type T4 of Acanthamoeba. This comparison further strengthens the hypothesis that Acanthamoeba sequence type T4 contains significant diversity equivalent to that which would occur separate several different species of other amoebae.
The tree below shows the neighbor-joining phylogeny for 18S rRNA gene sequences of the 32 isolates of V. vermiformis. There is little in the way of significant structure in genetic variation, given the close similarity of sequences. An attempt to find some patterns of variation was made by observing areas of the gene for which there appeared to be possible “allelic” variation between isolates. Ten small regions were identified, encompassing 38 variable positions in the gene. The allele identifier for each isolate is presented in the table below. Only two isolates share the complete set of “alleles” used in the analysis.
An arbitrary examination of the phylogenetic analysis suggests that there may be from four to seven sub-groups in the data. The “subtypes” are arbitrarily placed on the tree, but have only low levels of support, as seen from bootstrap values on the tree, reflecting the low level of genetic variation within these sequences. These “subtypes” are being used as the starting point for further studies (January 2017). (Groups 3a and 3b had previously been placed together in an earlier iteration of the figure.) Additional analysis of the partial sequences that make up ~95% of the sequences in the DNA database for V. vermiformis will provide further evidence of any genetic structure within V. vermiformis.
ALLELIC VARIATION WITHIN V. VERMIFORMIS
Isolates of V. vermiformis can also be classified by specific alleleic variation that characterizes a sequence. The alleles that we have identified correspond to a single code representing variation at a set of 36 single nucleotide polymorphisms that are spread over the length of the sequence of the 18S rRNA gene.
The allelic identification are given below for the sequences from 35 isolates, shown in the tree above, assigned to V. vermiformis . Nucleotide positions that showed a difference in a single isolate were not included (except where these sites fell within one of the variable segments). Additional positions may be added in the future as potential allelic information from partial sequences is better integrated into the analysis. Sites that are are scored “0” in the following table occur when a sequence in the database did not overlap with a particular portion of the gene.
Table 1. (updated December 2017)
Among the 30 isolates in Table 1 that show complete overlap of the variable regions, 14 represent unique (singleton) occurrences of an allele. Five alleles occur in more than one isolate. Three alleles occur twice. Two alleles occur three times. One allele (21112211122) occurs seven times.
SEQUENCE INFORMATION CONCERNING VARIABLE SEGMENTS IN TABLE 1:
Sites A through J in the table above represent the following segments and variation (in Bold/Italic) within the sequences of the V. vermiformis 18S rRNA genes of the isolates in the table.
SITE A: one variable nucleotide at position 71 in M95168 ; three alternative alleles
type 1 GC T AT
type 2 GC G AT
type 3 GC A AT
SITE B: one variable nucleotide at position 81 in M95168; two alleles
type 1 AC A GC
type 2 AC G GC
SITE C: one variable nucleotide at position 185 in M95168; two alleles
type 1 CC T GG
type 2 CC C GG
SITE D: two variable nucleotides (one an indel) at positions 284 and 286 in M95168; three alleles
type 1 TC – G T CG
type 2 TC G G T CG
type 3 TC G G A CG
SITE E5p: five variable nucleotides at positions 664, 667, 669, 670, 671 in M95168; ten alleles (revised December 2017)
type 1 TC T TT A G TGG TC
type 2 TC T TT A G CAG TC
type 3 TC C TT A G CAG TC
type 4 TC C TT A G CAA TC
type 5 TC C TT A G TAA TC
type 6 TC C TT T G CGG TC
type 7 TC C TT A G CGG TC
type 8 TC C TT A G TAG TC
type 9 TC C TT C G CGG TC
SITE E3p: four variable nucleotides at positions 693, 694, 695, and 697 in M95168; five alleles (revised December 2017)
type 1 GG TCA C T GG
type 2 GG TTG C T GG
type 3 GG CTG C T GG
type 4 GG TCG C A GG
type 5 GG TCG C T GG
SITE F: seven variable nucleotides (including two indels) at positions 724, 725, 726, 728, 730, 733 and 735 in M95168; nine alleles
type 1 TC AT – CC G C G AGG – G T GG
type 2 TC GT – CC G C G AGG – G C GG
type 3 TC AT – CC C C G AGG – G T GG
type 4 TC CCG CC G C G AGG C G G GG
type 5 TC AT – CC G C A AGG – G T GG
SITE G: one variable nucleotide (an indel) at position 1193 in M95168; two alleles
type 1 AA C CT
type 2 AA – CT
SITE H: three variable nucleotides at positions 1545, 1550 and 1551 in M95168; four alleles
type 1 GA C AGGG CT GG
type 2 GA T AGGG TT GG
type 3 GA T AGGG CC GG
type 4 GA T AGGG CT GG
SITE I: five variable nucleotides at positions 1628, 1650, 1658, 1663, and 1667 in M95168; five alleles
type 1 TA A GCGCGAGTCATCAACTCGCGC T GATTACG T CCCT G CCC T TT
type 2 TA G GCGCGAGTCATCAACTCGCGC C GATTACG T CCCT G CCC T TT
type 3 TA G GCGCGAGTCATCAACTCGCGC C GATTACG T CCCT A CCC T TT
type 4 TA A GCGCGAGTCATCAACTCGCGC T GATTACG T CCCT G CCC C TT
type 5 TA G GCGCGAGTCATCAACTCGCGC C GATTACG C CCCT G CCC T TT
SITE J: six variable nucleotides at positions 1730, 1731, 1732, 1734, 1750 and 1756 in M95168; six alleles
type 1 GG CAC G C AGGGGTCAAACCCTG T GTCCG T GC
type 2 GG CAC G C AGGGGTCAAACCCTG T GTCCG C GC
type 3 GG TAG G A AGGGGTCAAACCCTG T GTCCG T GC
type 4 GG CGC G C AGGGGTCAAACCCTG C GTCCG T GC
type 5 GG CGC G C AGGGGTCAAACCCTG T GTCCG T GC
type 6 GG GGC G C AGGGGTCAAACCCTG C GTCCG T GC