In collaboration with the lab of Rick Fishel, PhD, we are using two single molecule systems to study retroviral integration with recombinant proteins.
Total Internal Reflection Fluorescence (TIRF) Microscopy
 Using fluorescently labelled DNA or proteins we can observe single integration complexes interacting with target DNA in real time. These studies provide information about the length of each interaction event and the ability of retroviral integration complexes to slide or search target DNA.
Using fluorescently labelled DNA or proteins we can observe single integration complexes interacting with target DNA in real time. These studies provide information about the length of each interaction event and the ability of retroviral integration complexes to slide or search target DNA.
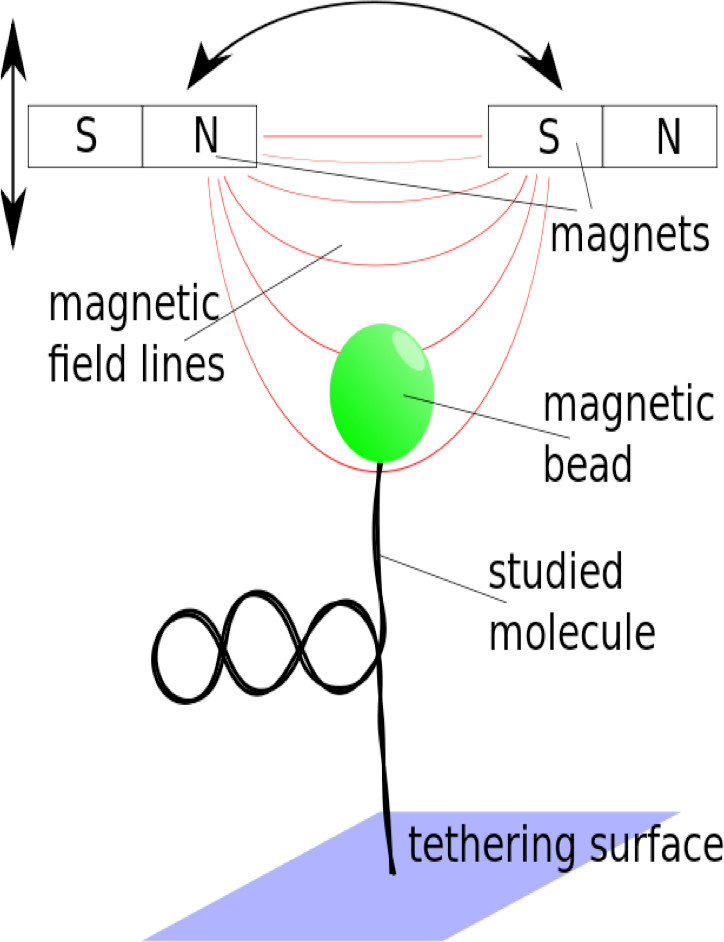
Magnetic Tweezers
A target DNA is linked to a surface and a magnetic bead. The focus of the bead indicates the height of the bead and information about the apparent length of the target DNA. The height of the bead may be altered by inducing supercoils in the target DNA or by integrating exogenous viral DNA.
PFV intasomes search DNA by 1D diffusion
Our initial studies of retroviral integrase by single molecule techniques focused on the prototype foamy virus (PFV) integrase. TIRF studies with purified PFV intasomes, a tetramer of integrase with Cy5 labeled viral DNA oligomers, revealed that the complexes may search 1 kilobase of naked DNA by 1 dimensional diffusion. In this kind of search, the PFV intasome complex is in constant contact with the DNA backbone; it moves like a nut on a screw rather than a washer. Magnetic tweezers experiments with PFV intasomes answered a long standing question in the field of retroviral integration: how much time between the joining of the two viral DNA ends? In the case of PFV integrase, the answer is less than half a second.