The Sullivan Lab collaborates with several expeditions to obtain diverse samples to answer important biological questions.
Global Oceanographic Research Expeditions
The Sullivan Lab is amassing an unprecedented archive of systematically-collected, metadata-rich ocean viral concentrates from two European-funded global oceanographic research expeditions, Tara Oceans and Malaspina. In total, we anticipate > 2,000 samples from multiple depths at 550 sites around the world’s oceans. Both expeditions explore the biology, chemistry, and physics of the oceans through large, international consortiums that include unprecedentedly diverse expertise. Further, both consortiums are built upon open exchange of ideas, well–documented voucher collections, and robust databasing to support sample archives. This foundation for diversity studies on a scale not previously possible allows us to explore the diversity, phylogeography, and genomic variability of ocean viral groups.
Tara Oceans
Tara Expeditions, a French non–profit organization active since 2003, organizes voyages to study the world’s oceans and the impact of climate change. Tara, a boat built for extreme conditions, is the platform for high–level scientific research missions around the globe.
Tara Oceans, a three year expedition that took place from September 2009 to November 2012, was the first attempt at a global study of marine plankton (viruses, bacteria, protists, and small metazoans). This expedition consisted of 375 stations sampling at 1 to 5 depths per station, to 1000m, mostly of the photic zone. Samples and data collected from across the world’s ocean environments are being analyzed by a scientific consortium of about 200 scientists with participation from 50 labs in 15 countries (including the Tucson Marine Phage Lab) to gain better knowledge of marine ecosystems. What do communities look like? How do various organisms interact with each other? How do they interact with their physico-chemical environment?
Malaspina
Malaspina Expedition 2010 is an interdisciplinary research project led by the Spanish National Research Council took place from December 2010 to July 2011. This expedition, aboard the ship Hesperides, voyaged through Rio de Janeiro, Cape Town, Perth, Sydney, Auckland, Honolulu, Cartagena of Indias, and Panama before returning to Spain. It sampled at 180 stations with 9 depths per station, to 5000m, of the photic zone and deep sea. About 300 scientists are directly involved with participation from 30 labs in 14 countries (including Tucson Marine Phage Lab). The goal of this expedition is to generate an inventory of the impact of global change on the ecosystems of the ocean and to explore their biodiversity, particularly in the deep ocean.
• Malaspina Expedition Website
Coastal to Open Ocean Transects
Line P
The LineP cruises from Vancouver to Ocean Station Papa traverse some 2000km along the climate-sensitive Oxygen Minimum Zone (OMZ) of the subarctic Eastern Northern Pacific Ocean. The subarctic Eastern North Pacific (ENP) Ocean hosts a massive, deep-water OMZ and large annual open-ocean dimethylsulfide (DMS) pulses. Marine OMZs are sources for climatologically active trace gases including methane, nitrous oxide, and dimethylsulfide, and major sinks for nitrogen. Microbially–mediated biological activity within these systems thus directly impacts both ocean productivity and global climate balance. In addition, ENP surface waters see large annual blooms of phytoplankton and bloom-associated bacteria that generate significant fluxes of DMS, which when “leaked” to the atmosphere influences global climate through cloud formation. The LineP transect of the ENP OMZ has been oceanographically monitored for >50 years, and in recent years has adopted a microbiological research focus.
Our research seeks to explore the roles that viruses play in these systems, in the context of Dr. Steve Hallam Lab’s microbial prokaryote work. What types of viruses are there, and how are they distributed throughout 4,000m of the water column? What microbial metabolisms might they be manipulating (e.g., photosynthesis)? How do their communities change over space and time?
Line 67
The Line67 transect was initiated ∼1950 as part of the CalCOFI program and is a designated time series site with over 20 years of contextual data maintained between MBARI and Scripps Institution of Oceanography. This coastal to open ocean transect traverses a Monterey Bay coastal area influenced by agricultural runoff, natural mesotrophic waters, and oligotrophic waters all through one of the most well–known and well–studied upwelling regions in the world, the Monterey Bay.
Our research focuses on studying how viral communities change along this oceanographically rich transect, as well as how viruses impact their microbial prokaryote and picoeukaryote hosts. This work is in collaboration with Dr. Alex Worden, MBARI.
Time Series
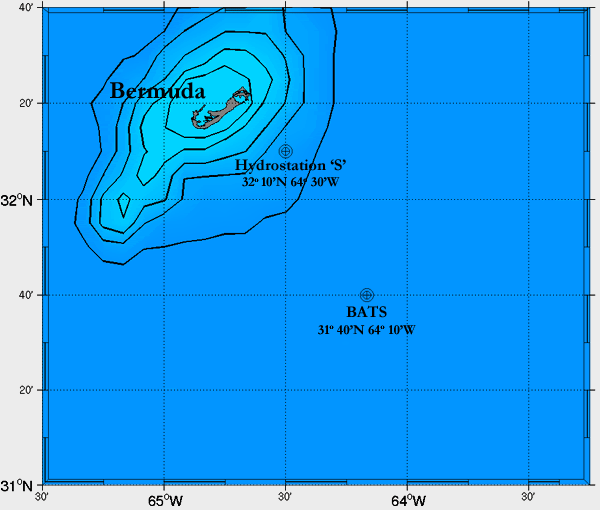
BATS (Bermuda Atlantic Time-series Study)
The Bermuda Atlantic Time-series Study (BATS) off the coast of Bermuda was established in 1988 to allow scientists to monitor data on dissolved carbon in the ocean and carbon dioxide in the atmosphere. This data helps scientists better understand global climate change and the ocean’s responses to variations in the Earth’s atmosphere, and can be used to study many ocean dynamics such as temporal variations in nutrient concentrations of surface water, phytoplankton blooms, the role of dissolved organic carbon in the global carbon cycle, and responses of calcifying organisms (like corals) to ocean acidification.
Here at the Sullivan Lab, we are partipating in the BATS sampling expeditions to understand how nutrient availability affects viral dynamics, the contributions of viral genes to nutrient uptake by the hosts during infection, and the chemical composition of nutrients released from host cells during viral infection.
Unique Environments
Permafrost
One third of the world’s carbon is held in permafrost soils; however, due to warmer temperatures, many permafrost regions have begun to thaw causing massive amounts of organic material become available to microbes, which metabolize the nutrients and release carbon dioxide and methane (both greenhouse gases). Understanding this process is a challenge due to the complexity of carbon dynamics. Scientists at the University of Arizona (Dr. Virgina Rich, Dr. Scott Saleska, and Dr. Matt Sullivan) have teamed with scientists at the University of Queensland in Australia, University of Stockholm in Sweden, and Florida State University to understand this important environmental system. Sampling and measurements are being collected from Stordalen Mire in Abisko National Park in Sweden, just north of the Arctic Circle. A key challenge of this work is to integrate ecosystem-scale measurements with molecular-scale studies of microorganisms and viruses.
• UANews article, Sept. 23, 2013: How Soil Microbes Affect Global Climate
Biosphere 2
The Biosphere 2 Ocean offers TMPL an accessible (<1 hours drive from Tucson) and diverse aquatic field site for studying ‘wild’ microbial and viral populations. This proximity to such a site provides both the opportunity for temporal and spatial characterization of these communities, as well as naturally complex source waters for evaluation of new tools for interrogating ‘wild’ populations.
Deep Hypersaline Pools
Buried 3,000m deep beneath the eastern Mediterannean Sea are extreme hypersaline pools that are saturated with either 5M NaCl or 6M MgCl2). Remarkably, microbial life exists here and functions, and scientists at the Institute for Coastal Marine Environment (IAMC) in Italy have figured out how sample not only within these hypersaline pools, but also with incredible accuracy (10s of centimeters) across the *interface* between the deep seawater and hypersaline pool waters. We hope to develop methods to create viral concentrates and metagenomic libraries from these challenging samples.




Comments are closed, but trackbacks and pingbacks are open.