Our research focuses on the biogenesis of energy-transducing membranes, in particular those evolved in mitochondria and chloroplasts. Using molecular genetics and biochemical approaches, we are pursuing efforts in two areas: 1) assembly of mitochondrial complex I and 2) thiol-disulfide chemistry in the thylakoid lumen. Our experimental models of study are the green alga Chlamydomonas and the land plant Arabidopsis.
The assembly of Complex I, an open question in mitochondrial biology:
With one FMN molecule, 8 FeS clusters, and more than 40 subunits, mitochondrial Complex I (NADH-ubiquinone oxidoreductase) is the largest protein complex of the respiratory chain (Fig. 1). This multimeric proton-pumping enzyme is of dual genetic origin; only a few subunits are synthesized in the mitochondria while the majority of its constituents are imported from the cytosol. Complex I assembly is a poorly understood process and has received intense attention in the medical field because Complex I deficiencies are the most prevalent cause of mitochondrial diseases in humans (e.g. neuromuscular and cardiovascular diseases). The fact that about 60% of patients with Complex I defects carry no mutations in the subunits encoded by nuclear or mitochondrial genes suggests that mutations in yet-to-be-discovered assembly factors may account for Complex I deficiencies. The lack of an experimental model with facile genetics has limited the molecular dissection of Complex I assembly. In collaboration with Dr. Remacle (University of Liège, Belgium) our group has developed the green alga Chlamydomonas as a genetically-tractable system to discover novel Complex I assembly factors.

Fig. 1: Overview of the mitochondrial respiratory chain. Complex I (blue box) is the entry point of electron transfer in the respiratory chain. It is defined by a membrane arm harboring the quinol reducing site (QH2) and a soluble arm carrying an FMN molecule and 8 FeS clusters (yellow rectangles). The proton gradient generated at the level of complex I, III, and IV is used by ATP synthase for ATP synthesis ND (in and out) are alternative dehydrogenase and AOX is the alternative oxidase. These enzymes are only present in mitochondria of algae, plants, some fungi, and parasites.
Chlamydomonas is ideally suited to study Complex I because mutants lacking Complex I are viable but display a slow-growth phenotype in the dark (Fig. 2). To uncover loci controlling Complex I biogenesis, we performed mutagenesis and screened for nuclear mutants exhibiting slow growth in the dark. From a forward genetic screen, we isolated amc1 to amc13 mutants (for assembly of mitochondrial complex I), which result in no or reduced Complex I activity. Biochemical analyses reveal that amc mutants accumulate reduced levels of intact Complex I or fail to accumulate a mature enzyme. In some amc mutants, the accumulation of a soluble subcomplex indicates an arrest in the assembly process. The locus defined by the amc5/amc7 alleles was shown to correspond to a gene encoding a small subunit of the membrane arm of Complex I, while AMC9 encodes a subunit of the soluble arm of Complex I. Chlamydomonas can be utilized to delineate the consequences of patient mutations on complex I function, and pathogenic mutations linked to Complex I dysfunction in humans could be reconstructed using the amc mutants. Recently, AMC1 was described as a novel Complex I assembly factor, a finding that underscores the value of a genetically tractable system for unraveling the molecular basis of Complex I assembly. AMC1 is a low-complexity protein that we propose is controlling the expression of the mitochondrial gene nad4 encoding a subunit of the membrane arm of Complex I. We are currently working towards elucidating the biochemical activity and site of action of recently discovered AMC proteins in the assembly process of complex I.

Fig. 2: Growth phenotype of Chlamydomonas Complex I mutant. Complex I deficiency results in a slow-growth phenotype in the green alga Chlamydomonas.
Thiol-disulfide chemistry in the thylakoid lumen of the chloroplast:
Cysteine residues in proteins exist under the reduced formed (as thiols) or oxidized (as disulfide bonds) (Fig. 3). In the cell, cysteines are maintained reduced or oxidized via the operation of redox enzymes, which have been best studied in the bacterial periplasm. Until recently, there was no support for the operation of such enzymes in the thylakoid lumen of the chloroplast, which is believed to be of bacterial origin and can be considered as equivalent to the bacterial periplasm.

Fig. 3: Thiol-disulfide chemistry sulfhydryl oxidation leads to disulfide bond formation and disulfide bond reduction yields free thiols. These reactions are under the control of catalysts, which were best studied in the bacterial periplasm. Our group is interested in detailing the operation of such pathways in the thylakoid lumen, a compartment topologically related to the periplasm.
Our initial interest in the redox pathways operating in the thylakoid stemmed from our work on the assembly of mitochondrial and chloroplast cytochromes c which are electron carriers involved in respiration and photosynthesis. Cytochromes c are defined by a CXXCH heme binding site, where heme is attached covalently to the cysteines of the motif. The cysteines have to be reduced in order for the heme to be attached. Heme attachment to apocytochromes c in mitochondria and chloroplasts is catalyzed by different machineries. Our investigation of chloroplast cytochrome c assembly led to the discovery of two transmembrane redox pathways, which are required to reduce a disulfide at the CXXCH motif of apocytochromes c, prior to heme attachment in the thylakoid lumen (Fig. 4). One pathway is defined by the membrane protein CCDA and a thioredoxin-like protein CCS5, which are postulated to act sequentially in transferring reducing power across the thylakoid membrane. The other pathway controlled by CCS4 also operates in disulfide bond reduction but the mechanism of delivery of reducing power across the membrane remains obscure. LTO1 (lumen thiol-oxidoreductase 1), a protein controlling at the thylakoid membrane was recognized to operate in a disulfide forming pathway. One relevant target of action of LTO1 is a lumen-localized subunit of photosystem II, a multimeric enzyme involved in photosynthesis. The finding that disulfide bond formation is a catalyzed reaction in the lumen is an exciting finding as the importance of disulfide bond for the function of several luminal proteins has now become apparent. We are now addressing experimental questions about the identity of the relevant targets of action of the redox pathways operating in the thylakoid lumen and how they control plastid biogenesis.
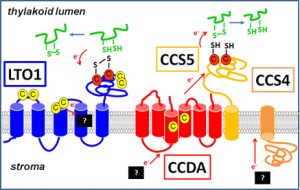
Fig. 4: Redox pathways in the lumen LTO1 defines the disulfide bond forming pathway. One disulfide bond reducing pathway is composed of CCDA and CCS5, while the other is controlled by CCS4. The electron acceptors and donors in the redox pathways are not known.
Hydrogen fuel production using green algae:
In collaboration with Dr. Hannah Shafaat (OSU) and Dr. Alexandra Dubini (University of Córdoba), my group took an interest in hydrogen (H2), a fossil fuel substitute that is naturally produced by several green algae including Chlamydomonas reinhardtii. Algal H2 is transiently produced by oxygen-sensitive hydrogenases via several pathways, two of which are linked to photosynthesis. To dissect the control of H2 production in the cell, we used a genetic approach and isolated a collection of ahp (attenuated for hydrogen production) mutants that are deficient for H2 evolution. We are also currently developing artificial metalloenzymes (rubredoxin) as an oxygen-tolerant substitute for the endogenous hydrogenases to design genetically engineered algal strains with enhanced H2 production capacity.