(revised October 2023)
Sequence type T5 was among the first sequence types identified. In contrast to other frequent sequence types such as T3, T4 or T11, isolates from sequence type T5 have almost all been assigned to a single taxa, Acanthamoeba lenticulata. Examination of the DNA sequences for the 18S rRNA gene in October 2023 shows that 482 isolates have been assigned into sequence type T5.
Sequence variability among the isolates assigned to sequence type T5 is much lower than that seen within T4. Because of this, we initally examined allelic variation within sequence type T5 using a different approach from that taken with other sequence types. Allelic variation is observed to be distributed primarily within two regions of the 18S rRNA gene of T5 isolates. One of these regions corresponds to a portion of stem-loop 19.1 of the Acanthamoeba 18S rRNA gene, the region showing hypervariation that is used to define alleles in T4 (and T3 and T11, as well) and which has been included in the partial sequence region designated DF3. This is a region that include nucleotides 1336 – 1363 of the sequence of A. lenticulata strain 45 (ATCC 50703; Genbank accession U94730). In T5, the length of this region is only about 50% the length of the variable regions in T3, T4 or T11. We designate this as “sub-allele region 2”. In addition there is a variable region of the gene from nucleotide 1096 – 1120 within the 18S rRNA gene of A. lenticulata strain 45 that we have identified as useful for allele classification and designate “sub-allele region 1”.
Ten different sub-allele region 1 sequences are found to occur in more than a single T5 isolate or uniquely in one of the 25 “almost-complete” T5 isolate sequence.
Sequences seen in allele region 1 that can be used to classify isolates of sequence type T5 include:
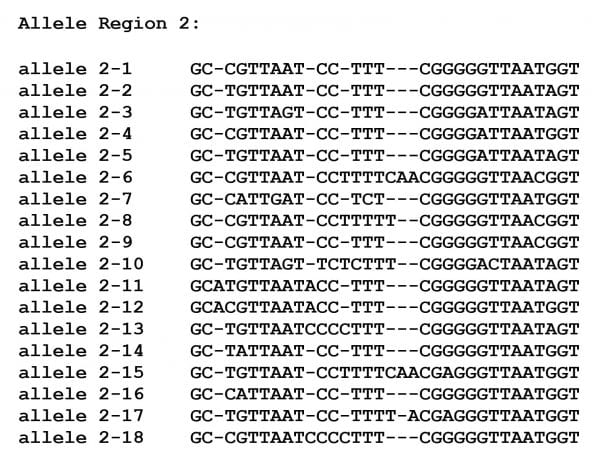
As of October 2023, there are 17 different sub-allele sequences seen in sub-allele region 2 that occurred in multiple isolates, together with a single additional allele that occurred in an almost complete 18S rRNA sequence:
When both regions are taken into account, isolates can be placed into a set of composite allele classes. In October 2023, 369 of the 482 T5 isolates could be classified into one of 20 composite allele classes that occur in multiple isolates in the dataset, while 11 isolates had combinations of shared alleles from the two regions that occurred only a single time. An additional 20 isolates carried a unique sub-allele at one or both of the sub-allele regions. The numbers in these composite allele classes that were shared by more than one isolate are shown in the following table (allele designation XX-YY, where XX = subregion 1 and YY = subregion 2:
Three of the composite allele classes account for almost 74% of the isolates that can be typed for both allele regions. It can be seen that substantial linkage disequilibrium exists for T5 isolates, with region 1 allele 1-1 is usually associated with either region 2 alleles 2-1 or 2-2, while region 1 allele 1-2 is usually associated with region 2 allele 2-3. This may be indicative of significant subdivision among the isolates that are placed within sequence type T5. It also suggests that genetic recombination between different copies of the 18S rRNA gene in Acanthamoeba is rare.
As with alleles in T3, T4 and T11, isolates of T5 that are not included in the tables above could have been excluded because they (a) did not have sequences that completely overlapped the regions of the two sub-alleles (n=67), (b) had sequence ambiguities that excluded them (n=8), (c) sequences that appeared to be a mixture from two isolates (n=5). As mentioned, there were some isolates for which the sequences of the 18S rRNA gene had a unique sequence combination for each of the two sub-regions amd represented combinations that were seen only once in the data (n=20).
Because two separate sequence regions were included in the composite alleles, including a region that was not homologous to that analyzed in the other sequence types, an additional analysis has also been performed.
ALLELES IN SUB-REGION 2
Sub-allele region 2 corresponds directly to the same hypervariable regions considered for Sequence types T3, T4 and T11. We have thus determined the frequency of isolates carrying specific alleles for sub-allele region 2. Again, it must be emphasized that region 2 is about 50% smaller than the equivalent DNA region within sequence type T3, T4 or T11. There are 417 T5 isolates that include sequences in sub-allele region 2. Of these 397 carry alleles for sub-region 2 that occur in more than a single isolate. The remaining isolates include 20 isolates with unique sequences in the subregion, 53 isolates that do not overlap the subregion and 12 isolates with amioguous or mixed sequences. The frequencies of the alleles that occur in more than one isolate are shown in the table below:
Three alleles dominate, region 2 alleles 2-1, 2-2 and 2-3. Together they represent 80% of the T5 isolates that have been typed for the DF3 region.